library(here)here() starts at /Users/kellymccormickhatfield/Documents/MADA 2023/kellyhatfield-MADA-portfoliolibrary(tidyverse)── Attaching packages
───────────────────────────────────────
tidyverse 1.3.2 ──✔ ggplot2 3.4.0 ✔ purrr 1.0.1
✔ tibble 3.1.8 ✔ dplyr 1.0.10
✔ tidyr 1.3.0 ✔ stringr 1.5.0
✔ readr 2.1.3 ✔ forcats 0.5.2
── Conflicts ────────────────────────────────────────── tidyverse_conflicts() ──
✖ dplyr::filter() masks stats::filter()
✖ dplyr::lag() masks stats::lag()library(tidymodels)── Attaching packages ────────────────────────────────────── tidymodels 1.0.0 ──
✔ broom 1.0.2 ✔ rsample 1.1.1
✔ dials 1.1.0 ✔ tune 1.0.1
✔ infer 1.0.4 ✔ workflows 1.1.2
✔ modeldata 1.0.1 ✔ workflowsets 1.0.0
✔ parsnip 1.0.3 ✔ yardstick 1.1.0
✔ recipes 1.0.4
── Conflicts ───────────────────────────────────────── tidymodels_conflicts() ──
✖ scales::discard() masks purrr::discard()
✖ dplyr::filter() masks stats::filter()
✖ recipes::fixed() masks stringr::fixed()
✖ dplyr::lag() masks stats::lag()
✖ yardstick::spec() masks readr::spec()
✖ recipes::step() masks stats::step()
• Learn how to get started at https://www.tidymodels.org/start/library(performance)
Attaching package: 'performance'
The following objects are masked from 'package:yardstick':
mae, rmseCleanSymp <- readRDS("~/Documents/MADA 2023/kellyhatfield-MADA-portfolio/fluanalysis/Data/CleanSymp.Rds")
ls(CleanSymp) [1] "AbPain" "BodyTemp" "Breathless"
[4] "ChestCongestion" "ChestPain" "ChillsSweats"
[7] "CoughIntensity" "CoughYN" "CoughYN2"
[10] "Diarrhea" "EarPn" "EyePn"
[13] "Fatigue" "Headache" "Hearing"
[16] "Insomnia" "ItchyEye" "Myalgia"
[19] "MyalgiaYN" "NasalCongestion" "Nausea"
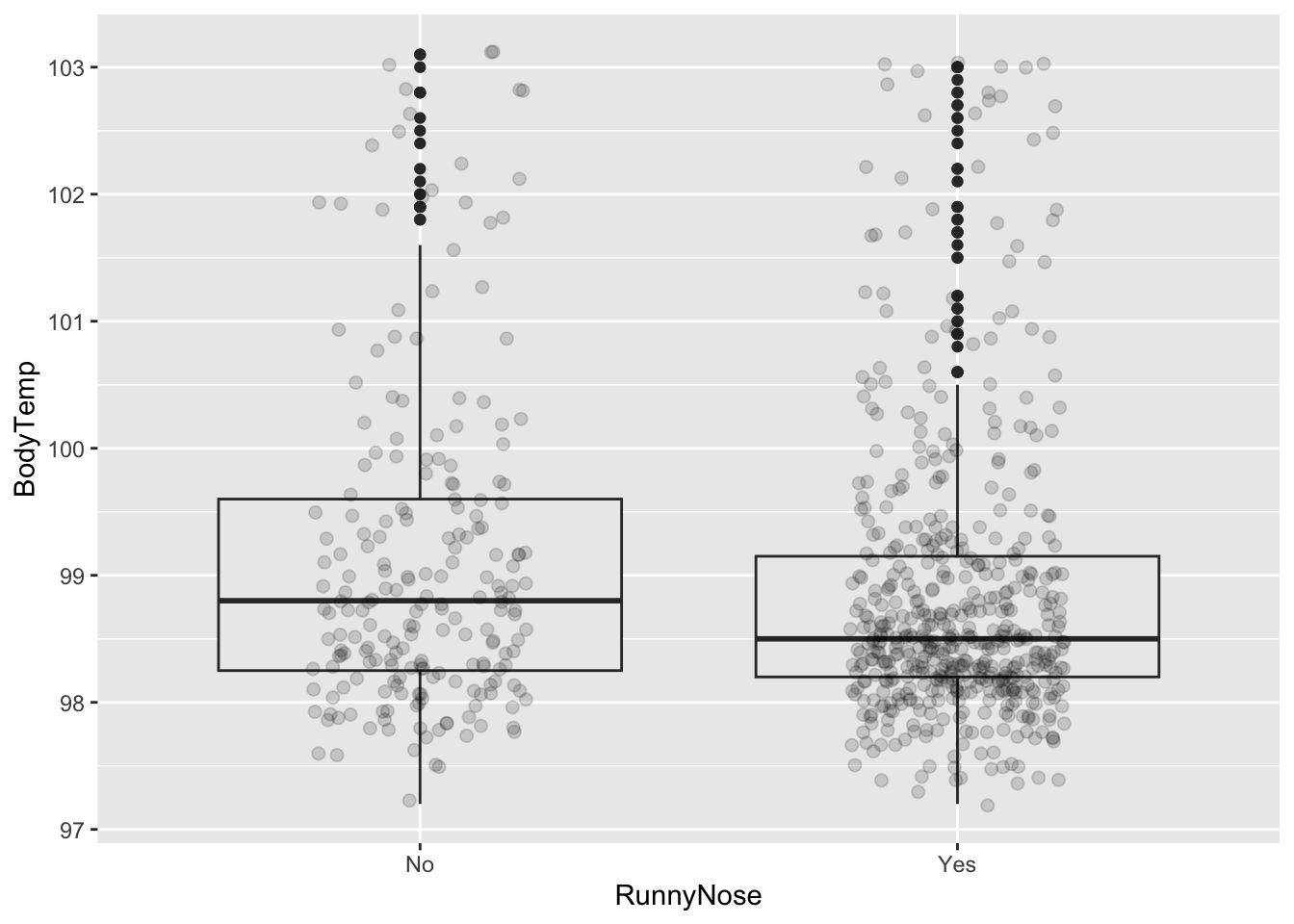
[22] "Pharyngitis" "RunnyNose" "Sneeze"
[25] "SubjectiveFever" "SwollenLymphNodes" "ToothPn"
[28] "Vision" "Vomit" "Weakness"
[31] "WeaknessYN" "Wheeze" summary(CleanSymp) SwollenLymphNodes ChestCongestion ChillsSweats NasalCongestion CoughYN
No :418 No :323 No :130 No :167 No : 75
Yes:312 Yes:407 Yes:600 Yes:563 Yes:655
Sneeze Fatigue SubjectiveFever Headache Weakness WeaknessYN
No :339 No : 64 No :230 No :115 None : 49 No : 49
Yes:391 Yes:666 Yes:500 Yes:615 Mild :223 Yes:681
Moderate:338
Severe :120
CoughIntensity CoughYN2 Myalgia MyalgiaYN RunnyNose AbPain
None : 47 No : 47 None : 79 No : 79 No :211 No :639
Mild :154 Yes:683 Mild :213 Yes:651 Yes:519 Yes: 91
Moderate:357 Moderate:325
Severe :172 Severe :113
ChestPain Diarrhea EyePn Insomnia ItchyEye Nausea EarPn
No :497 No :631 No :617 No :315 No :551 No :475 No :568
Yes:233 Yes: 99 Yes:113 Yes:415 Yes:179 Yes:255 Yes:162
Hearing Pharyngitis Breathless ToothPn Vision Vomit Wheeze
No :700 No :119 No :436 No :565 No :711 No :652 No :510
Yes: 30 Yes:611 Yes:294 Yes:165 Yes: 19 Yes: 78 Yes:220
BodyTemp
Min. : 97.20
1st Qu.: 98.20
Median : 98.50
Mean : 98.94
3rd Qu.: 99.30
Max. :103.10