Identify Variables
here() starts at /Users/kellymccormickhatfield/Documents/MADA 2023/kellyhatfield-MADA-portfolio
── Attaching packages
───────────────────────────────────────
tidyverse 1.3.2 ──
✔ ggplot2 3.4.0 ✔ purrr 1.0.1
✔ tibble 3.1.8 ✔ dplyr 1.0.10
✔ tidyr 1.3.0 ✔ stringr 1.5.0
✔ readr 2.1.3 ✔ forcats 0.5.2
── Conflicts ────────────────────────────────────────── tidyverse_conflicts() ──
✖ dplyr::filter() masks stats::filter()
✖ dplyr::lag() masks stats::lag()
── Attaching packages ────────────────────────────────────── tidymodels 1.0.0 ──
✔ broom 1.0.2 ✔ rsample 1.1.1
✔ dials 1.1.0 ✔ tune 1.0.1
✔ infer 1.0.4 ✔ workflows 1.1.2
✔ modeldata 1.0.1 ✔ workflowsets 1.0.0
✔ parsnip 1.0.3 ✔ yardstick 1.1.0
✔ recipes 1.0.4
── Conflicts ───────────────────────────────────────── tidymodels_conflicts() ──
✖ scales::discard() masks purrr::discard()
✖ dplyr::filter() masks stats::filter()
✖ recipes::fixed() masks stringr::fixed()
✖ dplyr::lag() masks stats::lag()
✖ yardstick::spec() masks readr::spec()
✖ recipes::step() masks stats::step()
• Learn how to get started at https://www.tidymodels.org/start/
<- readRDS ("~/Documents/MADA 2023/kellyhatfield-MADA-portfolio/fluanalysis/Data/CleanSymp.Rds" )ls (CleanSymp)
[1] "AbPain" "BodyTemp" "Breathless"
[4] "ChestCongestion" "ChestPain" "ChillsSweats"
[7] "CoughIntensity" "CoughYN" "CoughYN2"
[10] "Diarrhea" "EarPn" "EyePn"
[13] "Fatigue" "Headache" "Hearing"
[16] "Insomnia" "ItchyEye" "Myalgia"
[19] "MyalgiaYN" "NasalCongestion" "Nausea"
[22] "Pharyngitis" "RunnyNose" "Sneeze"
[25] "SubjectiveFever" "SwollenLymphNodes" "ToothPn"
[28] "Vision" "Vomit" "Weakness"
[31] "WeaknessYN" "Wheeze"
SwollenLymphNodes ChestCongestion ChillsSweats NasalCongestion CoughYN
No :418 No :323 No :130 No :167 No : 75
Yes:312 Yes:407 Yes:600 Yes:563 Yes:655
Sneeze Fatigue SubjectiveFever Headache Weakness WeaknessYN
No :339 No : 64 No :230 No :115 None : 49 No : 49
Yes:391 Yes:666 Yes:500 Yes:615 Mild :223 Yes:681
Moderate:338
Severe :120
CoughIntensity CoughYN2 Myalgia MyalgiaYN RunnyNose AbPain
None : 47 No : 47 None : 79 No : 79 No :211 No :639
Mild :154 Yes:683 Mild :213 Yes:651 Yes:519 Yes: 91
Moderate:357 Moderate:325
Severe :172 Severe :113
ChestPain Diarrhea EyePn Insomnia ItchyEye Nausea EarPn
No :497 No :631 No :617 No :315 No :551 No :475 No :568
Yes:233 Yes: 99 Yes:113 Yes:415 Yes:179 Yes:255 Yes:162
Hearing Pharyngitis Breathless ToothPn Vision Vomit Wheeze
No :700 No :119 No :436 No :565 No :711 No :652 No :510
Yes: 30 Yes:611 Yes:294 Yes:165 Yes: 19 Yes: 78 Yes:220
BodyTemp
Min. : 97.20
1st Qu.: 98.20
Median : 98.50
Mean : 98.94
3rd Qu.: 99.30
Max. :103.10
Things to note:
Most variables are categorical Yes/No.
Temperature is continuous from 97.2 to 103.1
Weakness, CoughIntensity, and Myalgia are scored None, Mild, Moderate, Severe
MyalgiaYN, CoughYN2, and WeaknessYN are all Yes/No versions of their corresponding intensity variable (None, Mild, Moderate, Severe)
Data Explorations: Body Temperature
First, we want to look at temperature with a few key variables. We have selected cough, chest pain and wheeze.
%>% summarize (min= min (BodyTemp),mean= mean (BodyTemp), q1 = quantile (BodyTemp, 0.25 ), median = mean (BodyTemp), q3 = quantile (BodyTemp, 0.75 ), max= max (BodyTemp))
min mean q1 median q3 max
1 97.2 98.93507 98.2 98.93507 99.3 103.1
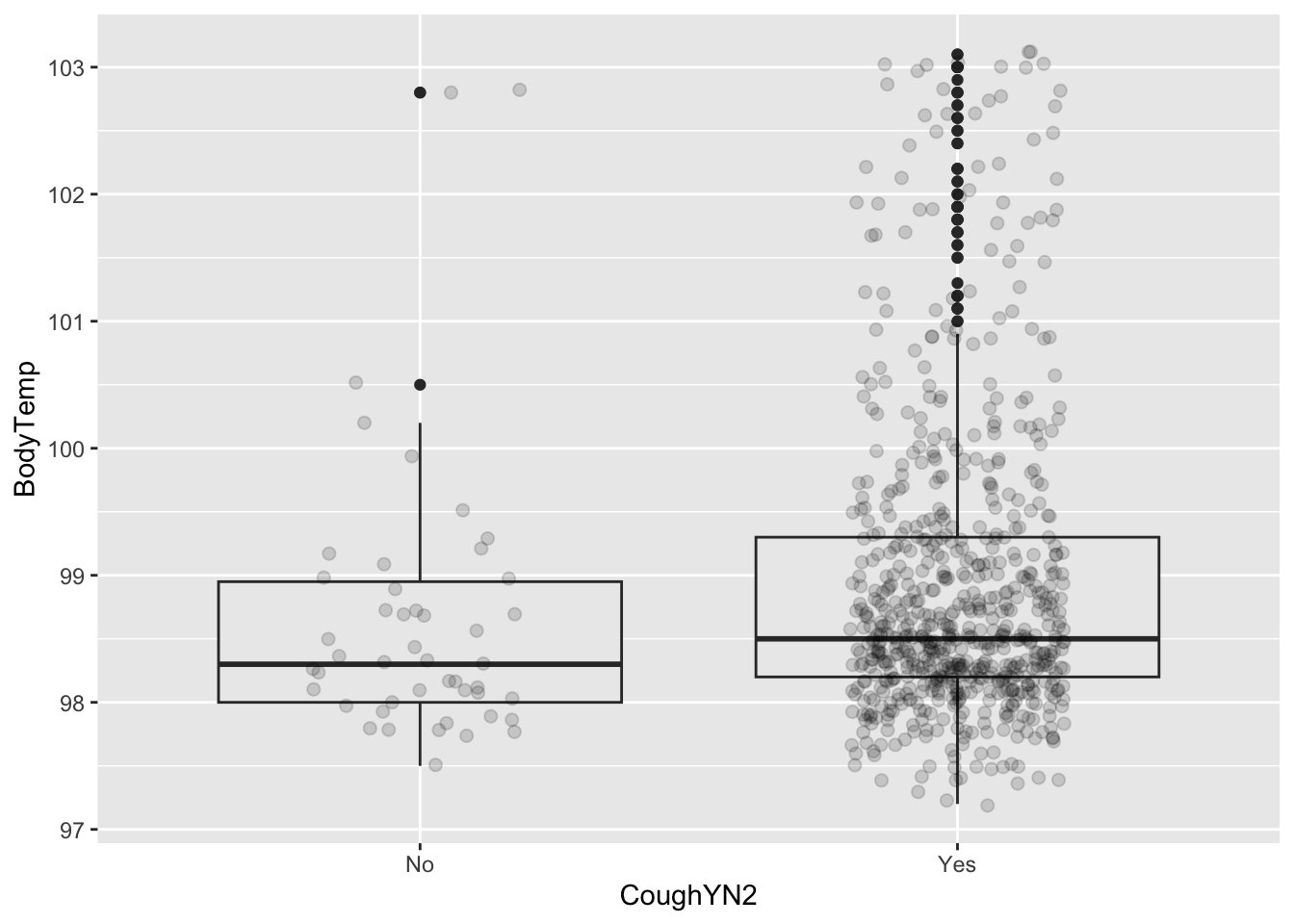
%>% group_by (CoughYN2) %>% summarize (mean= mean (BodyTemp), q1 = quantile (BodyTemp, 0.25 ), median = mean (BodyTemp), q3 = quantile (BodyTemp, 0.75 ))
# A tibble: 2 × 5
CoughYN2 mean q1 median q3
<fct> <dbl> <dbl> <dbl> <dbl>
1 No 98.7 98 98.7 99.0
2 Yes 99.0 98.2 99.0 99.3
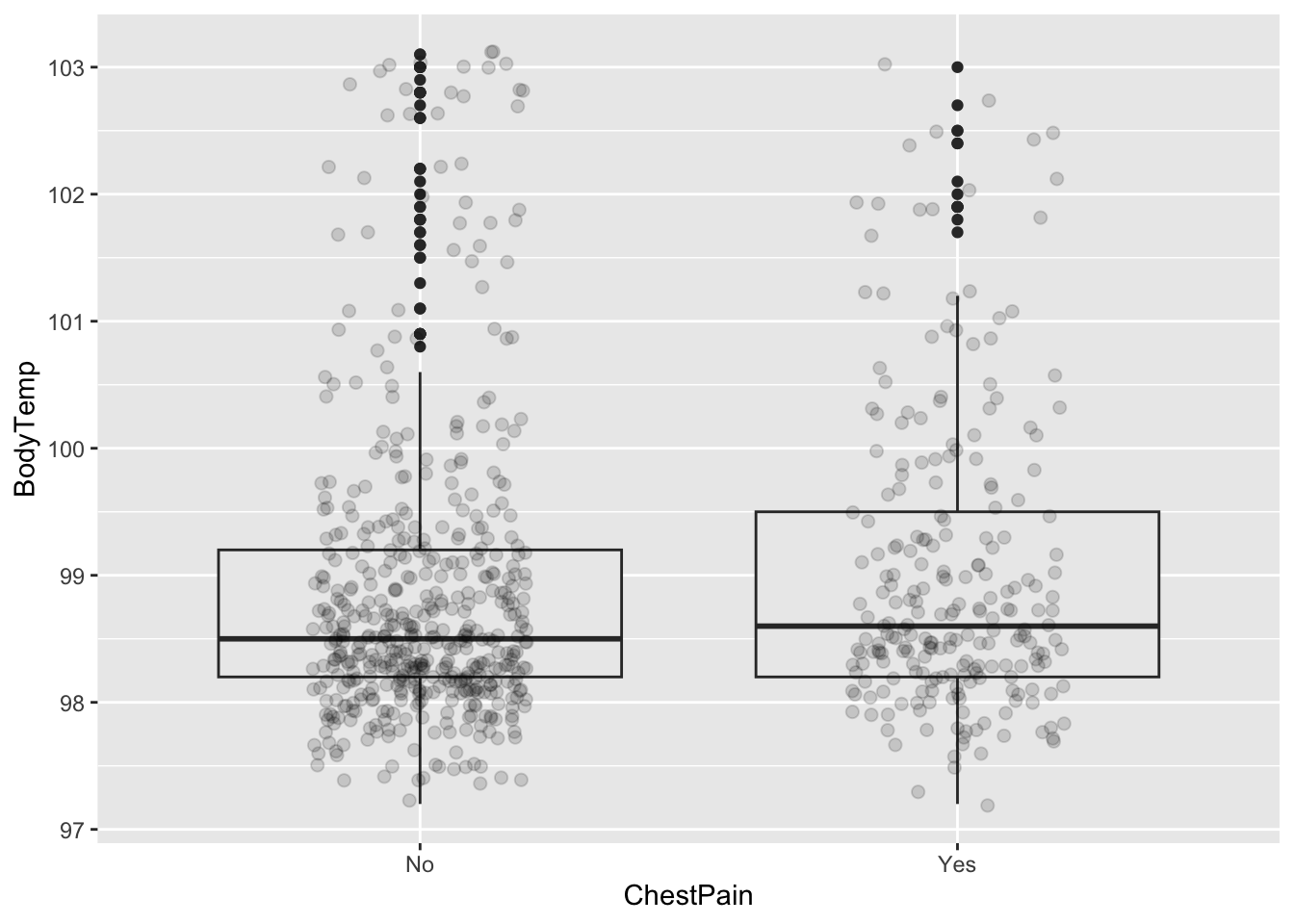
ggplot (CleanSymp, aes (x = CoughYN2, y = BodyTemp)) + geom_boxplot (fill = "grey92" ) + geom_point (size = 2 , alpha = .15 ,position = position_jitter (seed = 1 , width = .2 )) %>% group_by (ChestPain) %>% summarize (mean= mean (BodyTemp), q1 = quantile (BodyTemp, 0.25 ), median = median (BodyTemp), q3 = quantile (BodyTemp, 0.75 ))
# A tibble: 2 × 5
ChestPain mean q1 median q3
<fct> <dbl> <dbl> <dbl> <dbl>
1 No 98.9 98.2 98.5 99.2
2 Yes 99.0 98.2 98.6 99.5
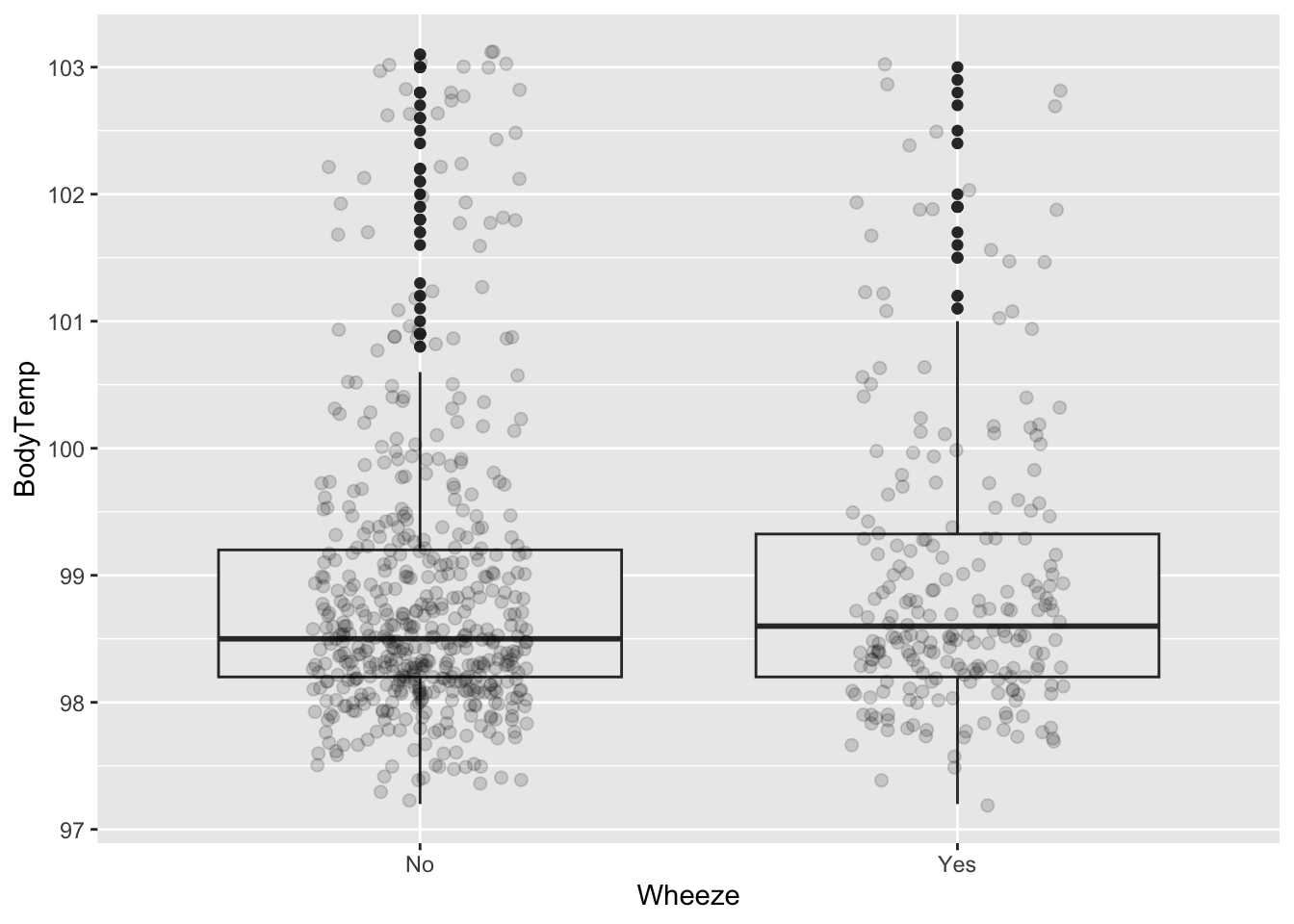
ggplot (CleanSymp, aes (x = ChestPain, y = BodyTemp)) + geom_boxplot (fill = "grey92" ) + geom_point (size = 2 , alpha = .15 ,position = position_jitter (seed = 1 , width = .2 )) %>% group_by (Wheeze) %>% summarize (mean= mean (BodyTemp), q1 = quantile (BodyTemp, 0.25 ), median = median (BodyTemp), q3 = quantile (BodyTemp, 0.75 ))
# A tibble: 2 × 5
Wheeze mean q1 median q3
<fct> <dbl> <dbl> <dbl> <dbl>
1 No 98.9 98.2 98.5 99.2
2 Yes 99.0 98.2 98.6 99.3
ggplot (CleanSymp, aes (x = Wheeze, y = BodyTemp)) + geom_boxplot (fill = "grey92" ) + geom_point (size = 2 , alpha = .15 ,position = position_jitter (seed = 1 , width = .2 ))
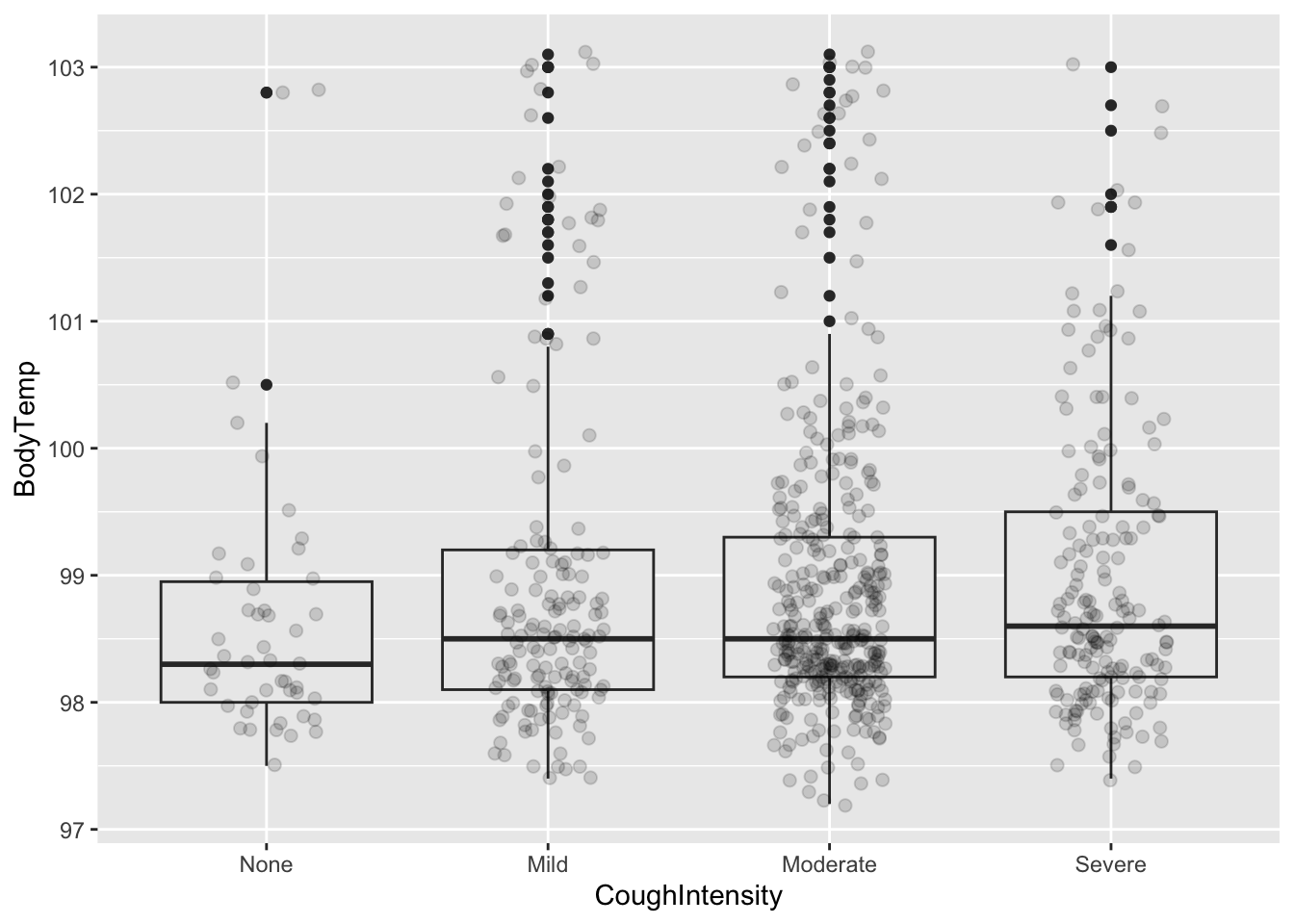
Since median teperature seems to be slightly elevated for the cough variable, we will look at it further for those varying rankings of the intensity of the cough.
%>% group_by (CoughIntensity) %>% summarize (mean= mean (BodyTemp), q1 = quantile (BodyTemp, 0.25 ), median = median (BodyTemp), q3 = quantile (BodyTemp, 0.75 ))
# A tibble: 4 × 5
CoughIntensity mean q1 median q3
<fct> <dbl> <dbl> <dbl> <dbl>
1 None 98.7 98 98.3 99.0
2 Mild 99 98.1 98.5 99.2
3 Moderate 98.9 98.2 98.5 99.3
4 Severe 99.0 98.2 98.6 99.5
ggplot (CleanSymp, aes (x = CoughIntensity, y = BodyTemp)) + geom_boxplot (fill = "grey92" ) + geom_point (size = 2 , alpha = .15 ,position = position_jitter (seed = 1 , width = .2 ))
The mean body temperature doesn’t seem to vary too much by increasing cough intensity group. However, perhaps some increase in median or q3 values.
Data Explorations: Nausea
For nausea we have decided to assess the relationship of nausea with subjective fever, myalgia, and abdominal pain.
# Variables of interest with Nausea <- table (CleanSymp$ Nausea,CleanSymp$ SubjectiveFever)
No Yes
No 166 309
Yes 64 191
prop.table (table1) %>% {.* 100 } %>% round (2 )
No Yes
No 22.74 42.33
Yes 8.77 26.16
<- table (CleanSymp$ Nausea,CleanSymp$ MyalgiaYN)
No Yes
No 63 412
Yes 16 239
prop.table (table2) %>% {.* 100 } %>% round (2 )
No Yes
No 8.63 56.44
Yes 2.19 32.74
<- table (CleanSymp$ Nausea,CleanSymp$ AbPain)
No Yes
No 444 31
Yes 195 60
prop.table (table3) %>% {.* 100 } %>% round (2 )
No Yes
No 60.82 4.25
Yes 26.71 8.22
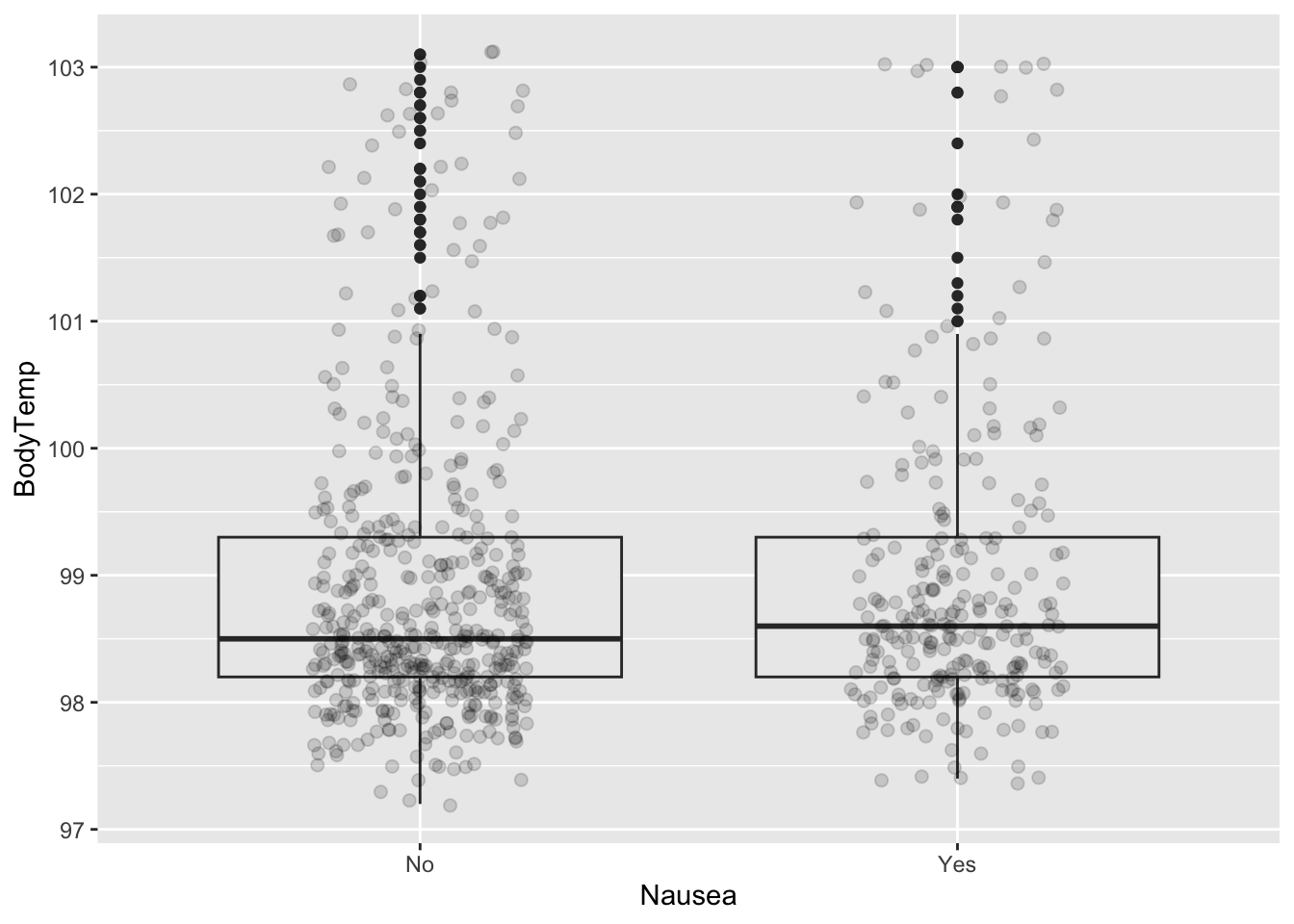
Data Explorations: Nausea and Body Temp
FFinally lets look at the relationships between our two primary variables.
%>% group_by (Nausea) %>% summarize (mean= mean (BodyTemp), q1 = quantile (BodyTemp, 0.25 ), median = median (BodyTemp), q3 = quantile (BodyTemp, 0.75 ))
# A tibble: 2 × 5
Nausea mean q1 median q3
<fct> <dbl> <dbl> <dbl> <dbl>
1 No 98.9 98.2 98.5 99.3
2 Yes 99.0 98.2 98.6 99.3
ggplot (CleanSymp, aes (x = Nausea, y = BodyTemp)) + geom_boxplot (fill = "grey92" ) + geom_point (size = 2 , alpha = .15 ,position = position_jitter (seed = 1 , width = .2 ))